Do fecal bile acid analyses allow taxonomic discrimination between Neotropical mustelids?
DOI:
https://doi.org/10.31687/SaremNMS23.6.4Keywords:
chemical profiles, Mustelidae, Neotropics, taxonomyAbstract
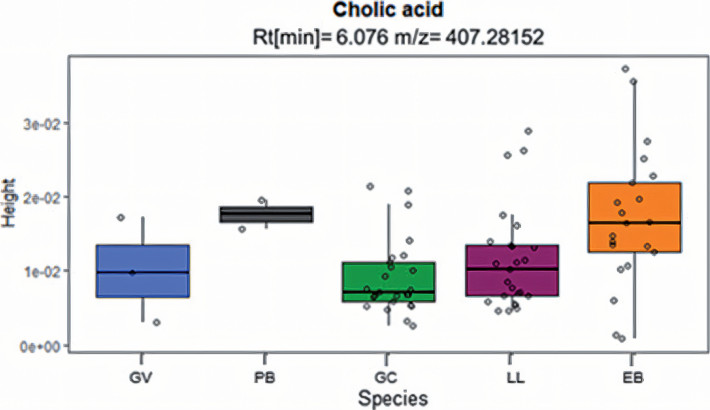
The use of fecal samples analysis to differentiate species is becoming more frequent in recent years. We performed a semi-quantitative fecal bile acid analysis of five Neotropical mustelids to access the diversity of bile acids, evaluating their taxonomic discriminatory potential. Fecal samples were collected from captive specimens and analyzed through liquid chromatography high-resolution mass spectrometry (LC-HRMS). We found significant differences between the analyzed species in four bile acids, supporting our method as a promising tool for taxonomic discrimination among mammals. Bile acids are endogenous compounds fundamental in biochemical pathways, so detecting interspecific differences may be relevant to understanding the physiology, health status and metabolic responses of mustelids to environmental changes.
References
Almeida, L. R., M. A. Alves, A. M. O. Mastella, R. Garrett, & M. J. R. Pereira. 2022. Neotropical mustelids: fecal metabolome diversity and its potential for taxonomic discrimination. Integrative Zoology 18:518–529. https://doi.org/10.1111/1749-4877.12645.
Alves, M. A., et al. 2021. A systematic pipeline to enhance the fecal metabolome coverage by LC-HRMS. Journal of the Brazilian Chemical Society 32:1435–1446.
Campbell-Palmer, R., & F. Rosell. 2011. The importance of chemical communication studies to mammalian conservation biology: a review. Biological Conservation 144:7:1919–1930. https://doi.org/10.1016/j.biocon.2011.04.028.
Araujo, M., M. Ciuccio, A. V. Cazon, & E. B. Casanave. 2010. Diferenciación de especies de Xenarthra (Mammalia) a través de la identificación de sus patrones de ácidos biliares fecales: una herramienta ecológica. Revista Chilena de Historia Natural 83:557–566. https://doi.org/10.4067/S0716-078X2010000400009.
Daruich, A., E. Picard, J. H. Boatright, & F. Behar-Cohen. 2019. Review: the bile acids urso- and tauroursodeoxycholic acid as neuroprotective therapies in retinal disease. Molecular Vision 25:610–624.
Guerrero , C., L. Espinoza, H. Niemeyer, & J. A. Simonetti. 2006. Using fecal profiles of bile acids to assess habitat use by threatened carnivores in the Maulino forest of central Chile. Revista Chilena de Historia Natural 79: 89–95. https://doi.org/10.4067/S0716-078X2006000100008.
Hagey, L. R., N. Vidal, A. F. Hofmann, & M. D. Krasowski. 2010. Evolutionary diversity of bile salts in reptiles and mammals, including analysis of ancient human and extinct giant ground sloth coprolites. BMC Evolutionary Biology 3:1–23. https://doi.org/10.1186/1471-2148-10-133.
Macdonald, D. W., C. Newman, & L. A. Harrington (eds.). 2017. Biology and conservation of Musteloids, 1st edition. Oxford University Press, New York. https://doi.org/10.1093/oso/9780198759805.003.001.
Mastella, A., C. Rodrigues, T. L. Kist, & M. J. Ramos Pereira. 2021. Take a good catch at the scat: carboxylic and sulfonic acid profiles as a non-invasive tool for species identification and sex determination in Neotropical carnivores. Studies on Neotropical Fauna and Environment. https://doi.org/10.1080/01650521.2021.1994786.
Matysik, S., C. I. Le Roy, G. Liebisch, & S. P. Claus. 2016. Metabolomics of fecal samples: a practical consideration. Trends in Food Science & Technology 57:244–255. https://doi.org/10.1016/j.tifs.2016.05.011.
Molinaro, A., A. Wahlström, & H.-U. Marschall. 2017. Role of bile acids in metabolic control. Trends in Endocrinology & Metabolism 29:31–41. https://doi.org/10.1016/j.tem.2017.11.002.
Nasini, U. B., N. Peddi, P. Ramidi, Y. Gartia, A. Ghosha, & A. U. Shaikh. 2013. Determination of bile acid profiles in scat samples of wild animals by liquid chromatography-electrospray mass spectrometry. Analytical Methods 5: 6319–6324. https://doi.org/10.1039/c3ay41048j.
RStudio Team. 2020. RStudio: integrated development for R. Boston, MA. <http://www.rstudio.com/>.
Salame-Méndez, A., et al. 2012. Método optimizado para evaluar ácidos biliares de muestras fecales secas o preservadas en etanol como herramienta para identificar carnívoros silvestres. Acta Zoologica Mexicana 28:305–320. https://doi.org/10.21829/azm.2012.282835.
Tsugawa, H., et al. 2015. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods 12:523–526. https://doi.org/10.1038/nmeth.3393.
Van den Berg , R. A., H. C. J. Hoefsloot, J. A. Westerhuis, A. K. Smilde, & M. J. van der Werf. 2006. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7:1–15. https://doi.org/10.1186/1471-2164-7-142.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Lana R. Almeida, Ana Maria O. Mastella, Marina A. Alves, Rafael Garret, Maria João R. Pereira

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.